-
Haploview Format File카테고리 없음 2020. 3. 1. 17:22

. 10 wrote:First of all, sorry for any inconvenience and thanks in advance for your consideration.
Haploview Format File Download

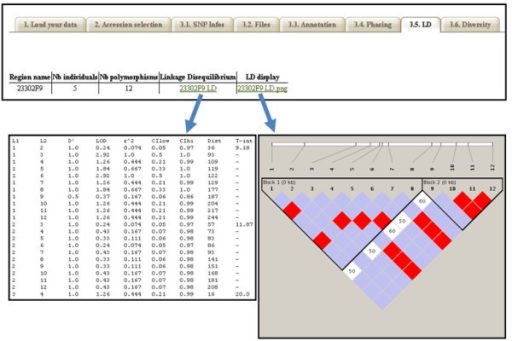
I am trying to analyse haplotype frequencies and associationsby using Haploview (it is the first time I use it), and I do not understand / know how to do it: I have 4 SNPs (C/T, C/A, C/G and AC). The Phased haplotypes input file format seems to be the easiest (at least for me):FAM1 FAM1M01 0 4 2 2FAM1 FAM1M01 0 4 2 2FAM1 FAM1F02 3 h 1 2FAM1 FAM1F02 3 h 1 2There are two lines (chromosomes) for each individual, 0 represents missing data, and h a heterozygous allele, but how can I assign the 1-4 numbers to the different genotypes?:1 CC CC CC AA, 2 CC AA CC AA, 3 CC CC GG AA, 4 CC CC CC CCAnd the rest of genotypes? (for example TT CC CC AA, TT AA GG CC, etc?)Thanks a lot!!jg.
Haploview Manual Pdf
Welcome to Haploview!!Introduction:Haploview is designed to simplify and expedite the process ofhaplotype analysis by providing a common interface to several tasksrelating to such analyses.What's here:A clean, pruned checkout of the code should have the following:edu/ - the source treeexamples/ - some sample input filesresources/ - some helper filesdocs/ - files related to program documentationbuild.xml - the ant build instruction fileREADME - this fileDependencies:HaploView can be built using the jakarta-ant build tool. Pleasedownload this tool at:Building:After installing ant and putting the ant executable script in your path:BASH export PATH=$PATH:CSH setenv PATH $PATH:simply type:ant haploviewQuestions??e-mail:haploview@broadinstitute.org.